News Archive
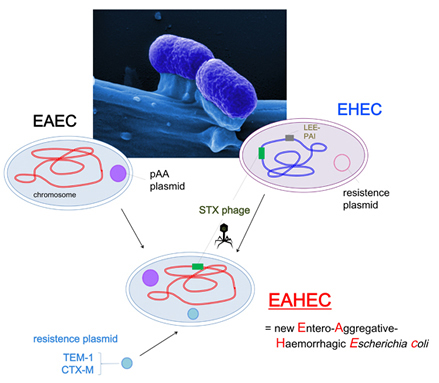
Not EHEC, but rather EAHEC (June 2011)
The sequence data indicate that the patient's isolates probably did not arise from an EHEC pathogen, but rather from a germ called EAEC (entero-aggregative Escherichia coli). This germ is able to attach particularly strongly to the epithelium. It then forms cell aggregates and initiates its normal, pathological programme. More than 96 percent of the genetic material of the Hamburg isolates are identical to that of an EAEC strain. The pathological potential of the EAEC germ has increased significantly by acquiring a special gene from other E. coli strains, such as EHEC, by means of bacterial viruses (phages) and integrating it into its own chromosome. This gene produces the so-called Shiga toxin, which originates from the pathogen causing shigellosis. It is a toxin which may trigger the haemolytic uraemic syndrome (HUS), namely blood decomposition, as well as complications, such as renal failure. This combination makes the bacterium dangerous: Its cells probably form large plaques in the intestine by attaching and aggregating, and this cell mass then produces a highly active poison. In addition, a so-called resistance plasmid protects the germ from a wide spectrum of antibiotics.
Scientists at the University of Göttingen have proposed the term ‘EAHEC’ (Entero-Aggregative-Haemorrhagic Escherichia coli) for the new pathotype.
Scientific publication:
Brzuszkiewicz E, Thürmer A, Schuldes J, Leimbach A, Liesegang H, Meyer F-D, Boelter J, Petersen H, Gottschalk G, and Daniel R (2011). Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC). Arch Microbiol 193:883-891 (abstract).
Modell of the formation of the new Entero-Aggregative Haemorrhagic Escherichia coli (EAHEC):